Spatial patterns of non-categorical rasters
Retrieve patterns from continuous data
Climate data - best practicse
First we need to setup our environment.
Using the geodata package (helper package for terra) we can download in a very comfortable often used core data sets. In this case Worldclim and
# German border on top level (nation)
de = geodata::gadm(country = "DEU",level = 0,path = tempdir())
# Worldclim historical climate data for Germany https://www.worldclim.org/data/index.html
wc_tmin_de = worldclim_country("Germany", var="tmin", path=tempdir())
# WCRP Coupled Model Intercomparison Project data https://www.wcrp-climate.org/wgcm-cmip
cmip_tmin_2_5 = geodata::cmip6_world("CNRM-CM6-1", "585", "2061-2080", var="tmin", res=10, path=tempdir())
# cropping and masking
wc_tmin_de = crop(wc_tmin_de,de,mask=T)
cmip_tmin_2_5_de = crop(cmip_tmin_2_5 , de,mask=T)
# set plotting layout
nf <- layout( matrix(c(1,2), ncol=2) )
plot(cmip_tmin_2_5_de)
plot(wc_tmin_de)Note: Please check the websites for the meaning of the data
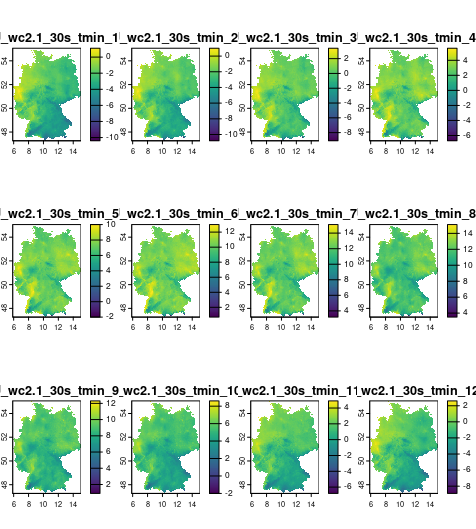
Minimum temperature (°C)
Identifying and comparing similar spatial patterns
Using the package spquery for finding similarities in continous raster data (e.g., raster time-series)
# get Marburg and make it a spatial point
mr = st_sf(st_sfc(st_point(c( 8.770833, 50.810001)),
crs = "EPSG:4326"))
vec = as.numeric(extract(wc_tmin_de, mr, ID = FALSE))
vec
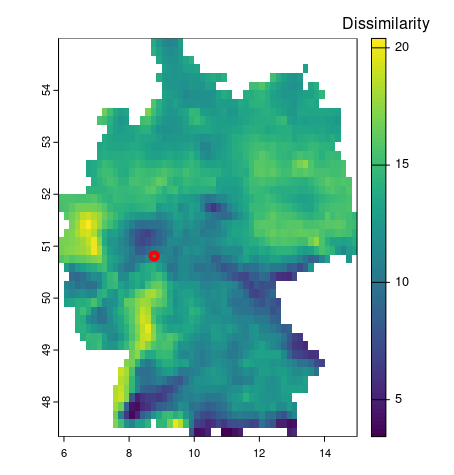
# search for similarity in the data set
search_tmin = spq_search(vec, cmip_tmin_2_5_de,
dist_fun = "euclidean")
# visualize
plot(search_tmin,
plg = list( loc = "topright",title = "Dissimilarity"))
plot(mr, add=TRUE, col='red', lwd=4)# some cleaning
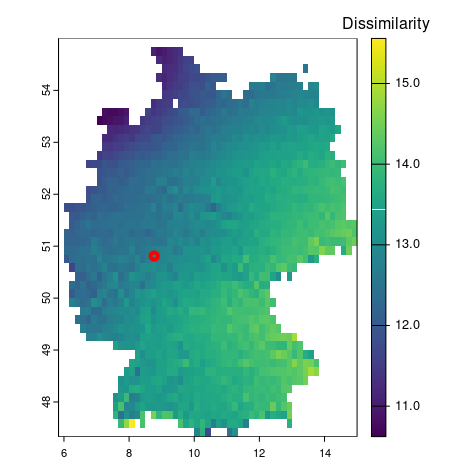
crop= crop( wc_tmin_de,cmip_tmin_2_5_de)
res = resample(crop,cmip_tmin_2_5_de)
# call comparison
compare_tmin = spq_compare(cmip_tmin_2_5_de, res,
dist_fun = "euclidean")
# visualize
plot(compare_tmin,
plg = list( loc = "topright",title = "Dissimilarity"),
col =viridis(256))
plot(mr, add=TRUE, col='red', lwd=4)patternogram
Describing the range of spatial autocorrelation
- explore spatial autocorrelations of predictors in machine learning models
- detect spatial autocorrelation in various data structures
- compare the spatial autocorrelation of variables over time
- investigate spatial autocorrelation of categorical spatial patterns
library(patternogram)
library(ggplot2)
pg1_100 = patternogram(wc_tmin_de, sample_size = 10)
pg2_100 = patternogram(cmip_tmin_2_5_de, sample_size = 10)
pg1_500 = patternogram(wc_tmin_de, sample_size = 100)
pg2_500 = patternogram(cmip_tmin_2_5_de, sample_size = 100)
pg1_1000 = patternogram(wc_tmin_de, sample_size = 1000)
pg2_1000 = patternogram(cmip_tmin_2_5_de, sample_size = 1000)
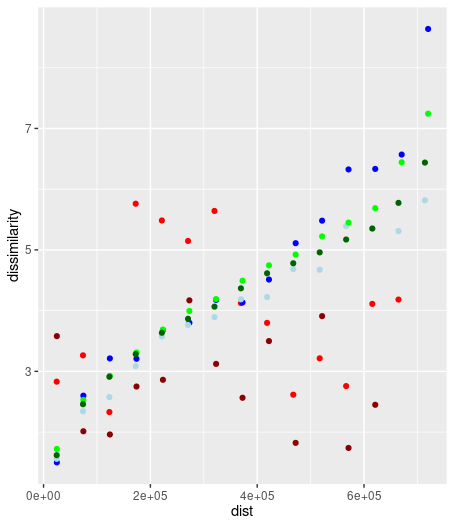
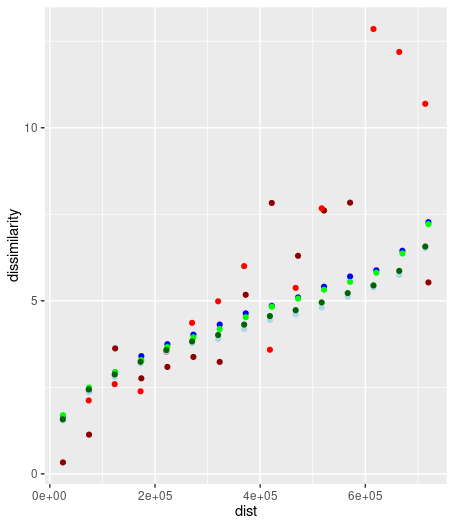
ggplot() + geom_point(data=pg1_100, aes(x=dist, y=dissimilarity), color='red4') +
geom_point(data=pg2_100, aes(x=dist, y=dissimilarity), color='red') +
geom_point(data=pg1_500, aes(x=dist, y=dissimilarity), color='blue') +
geom_point(data=pg2_500, aes(x=dist, y=dissimilarity), color='lightblue') +
geom_point(data=pg1_1000, aes(x=dist, y=dissimilarity), color='green') +
geom_point(data=pg2_1000, aes(x=dist, y=dissimilarity), color='darkgreen') library(patternogram)
library(ggplot2)
pg1_100 = patternogram(wc_tmin_de, width = 100, sample_size = 10)
pg2_100 = patternogram(cmip_tmin_2_5_de, width = 100 ,sample_size = 10)
pg1_500 = patternogram(wc_tmin_de, width = 100,sample_size = 10)
pg2_500 = patternogram(cmip_tmin_2_5_de, width = 100,sample_size = 10)
pg1_1000 = patternogram(wc_tmin_de, width = 100,sample_size = 10)
pg2_1000 = patternogram(cmip_tmin_2_5_de, width = 100,sample_size = 10)
ggplot() + geom_point(data=pg1_100, aes(x=dist, y=dissimilarity), color='red4') +
geom_point(data=pg2_100, aes(x=dist, y=dissimilarity), color='red') +
geom_point(data=pg1_500, aes(x=dist, y=dissimilarity), color='blue') +
geom_point(data=pg2_500, aes(x=dist, y=dissimilarity), color='lightblue') +
geom_point(data=pg1_1000, aes(x=dist, y=dissimilarity), color='green') +
geom_point(data=pg2_1000, aes(x=dist, y=dissimilarity), color='darkgreen') Slic /supercells
supercells: an extension of SLIC (Simple Linear Iterative Clustering; Achanta et al. (2012), doi:10.1109/TPAMI.2012.120) that can be applied to non-imagery geospatial rasters that carry:
- pattern information (co-occurrence matrices)
- compositional information (histograms)
- time-series information (ordered sequences)
- other forms of information for which the use of Euclidean distance may not be justified
- Segmentation/regionalization: partitioning space into smaller segments while minimizing internal inhomogeneity and maximizing external isolation
SLIC/supercells are a way to improve the output and reduce the cost of segmentation
Code examples
library(terra)
library(supercells)
# Version 1
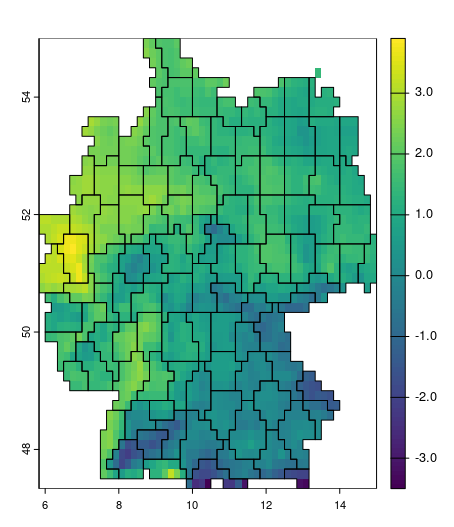
mintemp_zones = supercells(cmip_tmin_2_5_de, k = 250, compactness = 4)
plot(cmip_tmin_2_5_de[[1]]); plot(mintemp_zones, add = TRUE, col = NA)
# Version 2
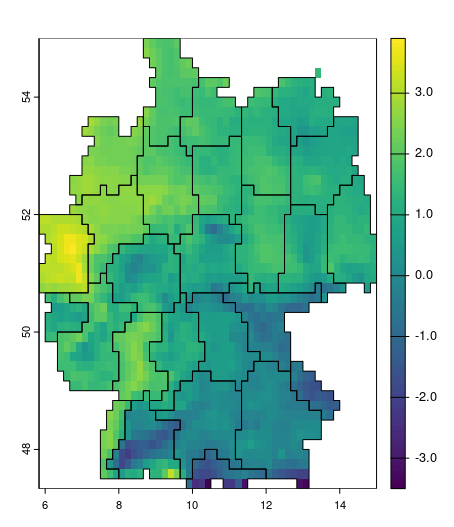
mintemp_zones = supercells(cmip_tmin_2_5_de, k = 50, compactness = 4)
plot(cmip_tmin_2_5_de[[1]]); plot(mintemp_zones, add = TRUE, col = NA)
# Version 3
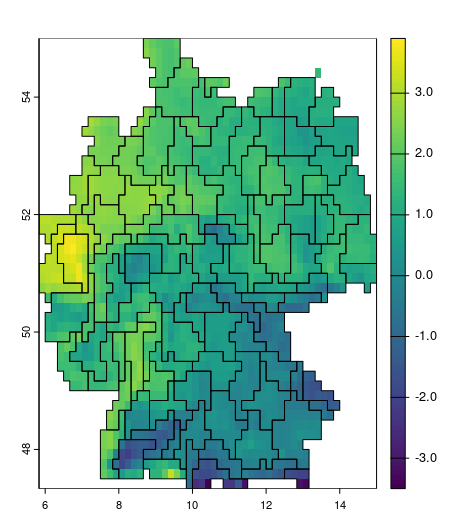
mintemp_zones = supercells(cmip_tmin_2_5_de, k = 250, compactness = 1)
plot(cmip_tmin_2_5_de[[1]]); plot(mintemp_zones, add = TRUE, col = NA)Code Examples - Helper functions
library(terra)
library(sf)
library(supercells)
library(rgeoda)
library(purrr)
library(tmap)
rawdata="supercells-examples-main/raw-data/"
# create helper functions -------------------------------------------------
get_dtw2d = function(x){
dist_mat = matrix(nrow = nrow(x), ncol = nrow(x))
for (i in seq_len(nrow(x))){
mat1 = matrix(unlist(x[i, ]), ncol = 2)
for (j in seq_len(nrow(x))){
mat2 = matrix(unlist(x[j, ]), ncol = 2)
dist_mat[i, j] = dtwclust::dtw_basic(mat1, mat2, norm = "L2", step.pattern = dtw::symmetric2)
}
}
stats::as.dist(dist_mat)
}
dtw_2d = function(x, y){
dtw2ddistance = dtwclust::dtw_basic(x = matrix(x, ncol = 2), y = matrix(y, ncol = 2),
norm = "L2", step.pattern = dtw::symmetric2,
error.check = FALSE)
return(dtw2ddistance)
}
regionalize_dtw_2d = function(k, superpixels, ...){
weight_df = st_drop_geometry(superpixels[, !colnames(superpixels) %in% c("supercells", "x", "y")])
weight_dist = get_dtw2d(weight_df)
rook_w = rook_weights(superpixels)
skater_results = ?skater(k, rook_w, weight_df, random_seed = 1, cpu_threads = 1, scale_method = "raw",
rdist = weight_dist)
superpixels$cluster = skater_results$Clusters
regions = aggregate(superpixels, by = list(superpixels$cluster), mean)
regions = st_cast(regions, "POLYGON")
regions$k = k
return(regions)
}
# normalize function
scale_01 = function(r){
# get the min max values
minmax_r = range(values(r), na.rm=TRUE)
# rescale
return( (r-minmax_r[1]) / (diff(minmax_r)))
}Code Examples - Regionalisation based on supercells
# create supercells based on the 2D time-series ---------------------------
sp = supercells(c(ta, pr), step = 15, compactness = 0.01, dist_fun = dtw_2d)
plot(sp)
# create 3, 7, 11, and 15 regions based on the 2D time-series -------------
sp_regions = ?map_dfr(c(3, 7, 11, 15), regionalize_dtw_2d, sp)
tm_shape(sp_regions) +
tm_polygons() +
tm_facets("k")Exercises
The goal: to regionalize Germany’s climates
- Use Worldclim versus CMIP data
- Use the upper helper functions and code for Great Britain
Extended SLIC workflow uses the dynamic time warping (DTW) distance function rather than the Euclidean distance.
<gisma 2024>
![CMIP6 downscaled future climate projection for 2061-2080 [model: CNRM-ESM2-1; ssp: “585”]](../assets/cmip.png)